Biochemist Swarts studies the immune system of viruses. Medema’s research focuses on the chemical language of our microbiome, the bacteria with which we coexist.
Immune systems
Bacteria are under constant attack by viruses. Thus, they have developed different immune systems, each capable of recognising specific virus particles. Some of these systems, such as the well-known CRISPR-Cas system, chop up harmful viruses or emit a warning signal to prompt other systems to neutralise the virus. These bacterial systems are used in testing for viruses, but also to cut and alter DNA.
Deviations
Because viruses constantly mutate to escape the immune systems, bacteria have a myriad of immune systems. Swarts searches for hitherto unknown systems in bacteria to discover new virus-identifiers. He also aims to discover whether they will allow him to identify and repair deviations in DNA.
Swarts intends to use the Starting Grant he was awarded by the European Research Council (ERC) to recruit three PhD students and an analyst. His team will use, among other techniques, X-ray crystallography to generate 3D images of the minute immune systems, in order to study how they identify viruses.
He may well discover immune systems that can be isolated and reprogrammes to rapidly identify a choice of viruses. Such a programmable virus test could prove very useful for testing large populations during a viral outbreak. Swarts has previously characterised an immune system that can now be used in the detection of COVID-19.
Chemical language
Bioinformatician Marnix Medema is searching for the chemical language our microbiome, the bacteria with which we coexist, communicates.
Humans, animals and plants coexist with billions of bacteria that are essential to their health, provide useful nutrients and protect against stress and disease. Recent studies have shown that the microbial communities are supported by specialised molecules that manage the communication between bacteria and with the bacteria’s host organism through a ‘chemical language’. These substances are generated by genes that are exchanged between different strains of bacteria.
Software
Medema wants to map this chemical language. He strives to discover what molecules are generated by which bacteria, and how these substances contribute to the health of man, animal and plant.

He intends to put the awarded ERC-grant to use to develop new bio-information software that can map this language and decode its meaning. He strives to identify the genes that regulate the exchange of information between the bacteria, and the patterns in which they are activated and deactivated. He also seeks information on the specific chemical structure of the molecules in question.
With the help of this data, he hopes to develop new algorithms capable of predicting what the molecules look like and what interactions they facilitate, both between bacteria and between bacteria and their host. This knowledge may make it possible to redesign bacterial communities to better protect crops from disease and drought, or to cure metabolic disorders in humans.

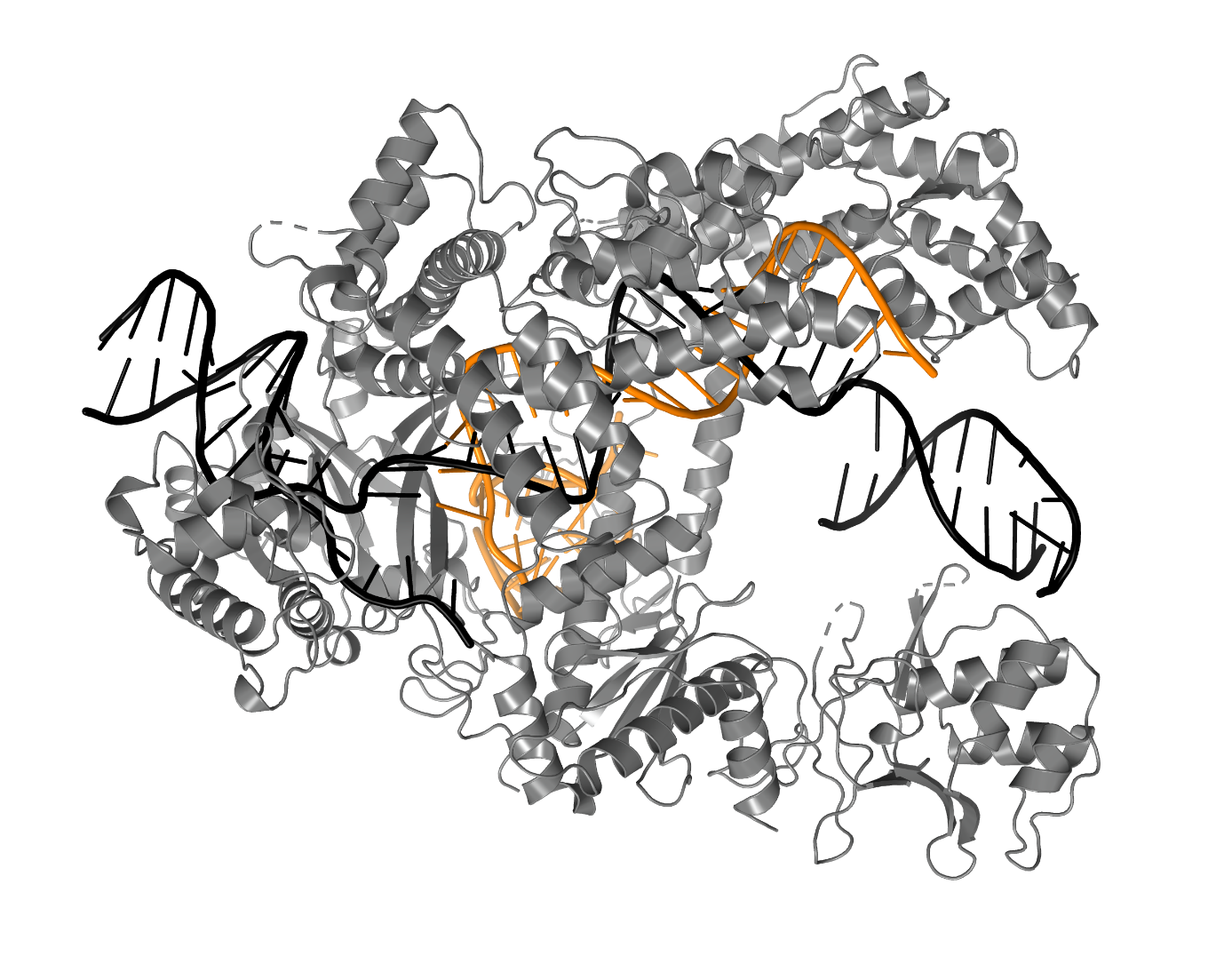 3D-impression of a bacterial immune system Image: Daan Swarts
3D-impression of a bacterial immune system Image: Daan Swarts